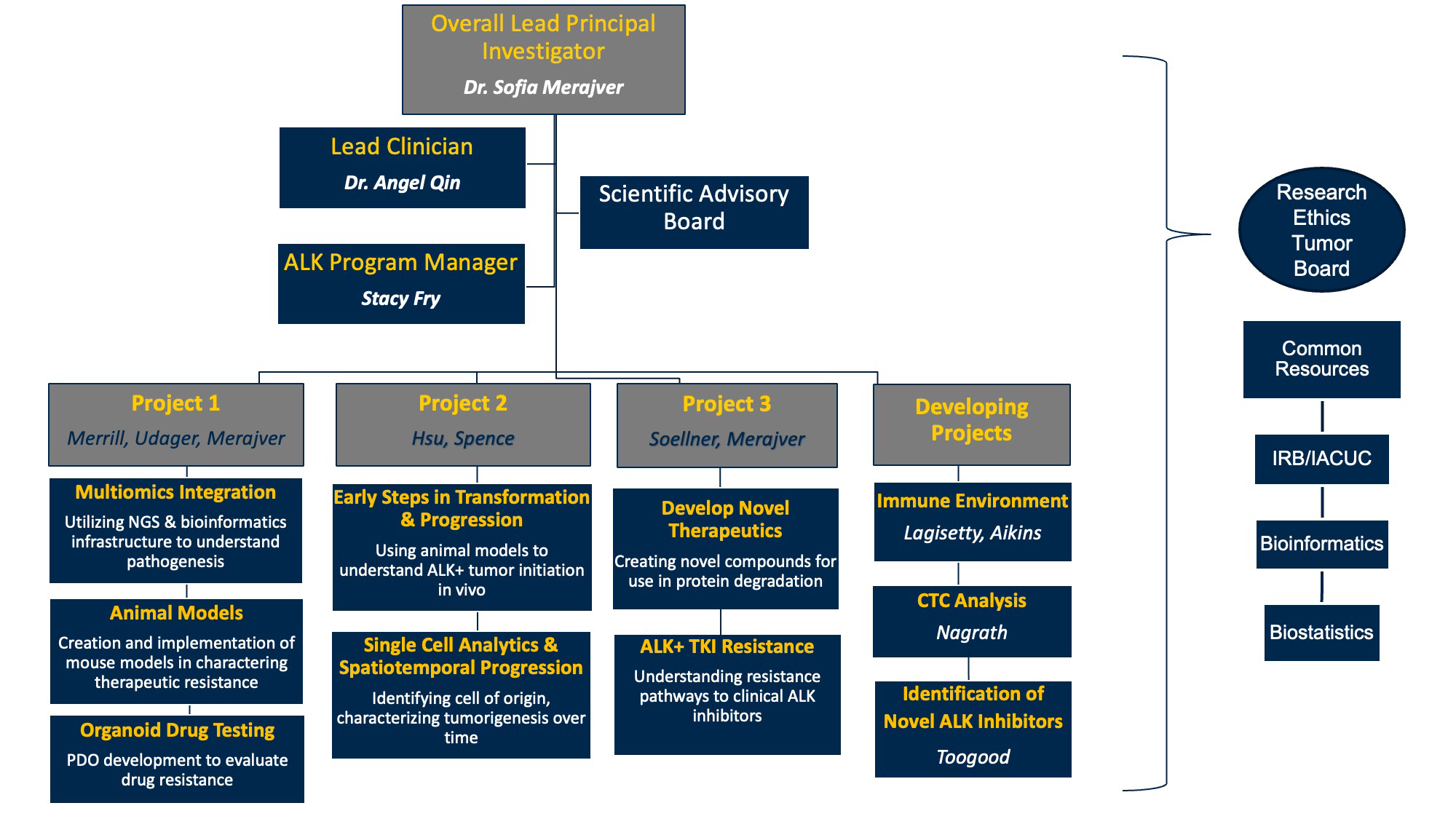
Research Projects Judith Tam ALK Lung Cancer Research Initiative
Project 1: Understanding efficacy and resistance in ALK NSCLC through tumor organoid drug testing and multi-omic integration
Project Co-Leaders:
- Sofia Merajver, M.D., Ph.D.
- Nathan Merrill, Ph.D.
- Aaron Udager, M.D., Ph.D.
Conventional cancer cell lines and in vitro models can be useful but are often limited as they do not maintain the complex tumoral and cell-type heterogeneity that occurs in the patient. Moreover, generation of these cell lines and laboratory models often occurs on a timescale that cannot immediately impact patient treatment decisions. Although TKIs are an excellent treatment for oncogene-driven lung cancer, after TKIs are exhausted, next line therapy options are limited and are chosen empirically without any guidance from the actual tumor. We can improve on this situation for lung cancer in a very short time because we have already developed all the methods needed to accomplish this task. Based on a currently existing platform at the University of Michigan that has successfully profiled lung cancers metastatic to brain, breast cancer and bladder cancer tissues directly from patients at biopsy or surgery, we seek to perform sensitivity profiling with a panel of 30-50 or more drugs and innovative drug combinations, using fresh tumor tissue within 5-7 days after tissue resection. As the samples will be minimally cultured, we expect that the profiling will more accurately reflect the responses of the tumors as they would occur in the patient. We currently have access to surgical tissue from lung cancer resections but propose to expand our tissue procurement pipeline to include samples obtained from interventional radiology, and at other centers nationally and internationally, thereby allowing us to test tissue from metastatic sites and sites of recurrence or progression from many diverse patients. The profiling will occur within 1-2 weeks, and therefore once certified, would allow for rapid actionable information which could directly inform which therapies are received by the patient under clinical protocols.
Project Objectives:
- Objective 1: Test minimally cultured non-small cell lung cancer samples, obtained at the time of diagnosis and recurrence or progression.
- Objective 2: Molecularly characterize PDOs to identify therapeutic vulnerabilities and changes over the course of pathogenesis.
- Objective 3: Utilize NGS and bioinformatics infrastructure to characterize pathogenesis.
Project 2: Modeling and characterizing early ALK-dependent tumorigenesis
Project Co-Leaders:
- Peggy Hsu, M.D., Ph.D.
- Jason Spence, Ph.D.
ALK+ lung cancer is often treatable but incurable at time of diagnosis. The short-term goal of this project is to characterize the earliest cellular and molecular changes that occur in the lung epithelium upon ALK mutation with the longer-term goal to use these insights to guide strategies for early detection, intervention, disruption of cancer progression, and ultimately to cure patients with ALK+ lung cancer. Limited access to early-stage ALK+ patient samples has prevented its study. The reason for this is two-fold: (1) The disease is typically diagnosed at an advanced stage after metastasis has already occurred. (2) Sequencing of cancer-causing mutations is not performed for early-stage lung cancer given that it does not generally change clinical management. Through the Judith Tam ALK Lung Cancer Research Initiative, we have been able to access and analyze two rare early-stage ALK+ patient samples. However, there is a need to complement the data generated from these samples with alternative approaches. We are therefore modeling the process by which a normal stem cell acquires the ALK translocation both in stem cell cultures (organoids) and in the mouse to answer questions regarding cell of origin, the effect of ALK on cell state and plasticity, and the role of candidate downstream pathways in tumor initiation.
Project Objectives:
- Objective 1: Test the hypothesis that the cell of origin in human ALK+ lung cancer is the AT2 cell, and profile early effects of ALK activation on AT2 cell state.
- Objective 2: Model early ALK+ lung cancer in mouse organoids and mouse models.
- Objective 3: Identify and functionally interrogate relevant pathways downstream of ALK in early lung cancer.
Project 3: Pursuing new approaches to invent and test a new strategy for using combinations of drugs against ALK and create novel inhibitors against ALK, by targeting a portion of the ALK molecule not usually targeted by conventional therapies
Project Co-Leaders:
- Sofia Merajver, M.D., Ph.D.
- Matthew Soellner, Ph.D.
Drug combinations are commonly employed in cancer therapeutics to increase anti-tumor efficacy and decrease the rates of resistance. In chronic myelogenous leukemia, for examples, five FDA-approved drugs address a specific genetic challenge, with each taking on a different resistance mechanism. Recently, clinical trials have shown that combinations of these approved inhibitors can improve outcomes. To date, this strategy has not been explored with ALK rearrangement in non-small cell lung cancer. Here, we propose evaluating the combination of ALK inhibitors in patient samples to determine whether the combinations, such as the leukemia example, lead to reduced resistance and increased anti-tumor efficacy. In addition to using the six FDA-approved ALK inhibitors, we will also undertake efforts to identify allosteric ALK inhibitors that bind and inhibit ALK kinase in a distinct location from the six approved inhibitors. Ultimately, we aim to identify optimal combinations of ALK inhibitors that will reduce resistance and increase anti-tumor efficacy to enhance progression-free survival in patients with ALK-rearranged non-small cell lung cancer. Other drug companions of ALK inhibitors can also be explored, based on these results.
Project Objectives:
- Objective 1: Understand resistance pathways to clinical ALK inhibitors. We propose to characterize a panel of EML4- ALK transformed non-small cell lung cancer cell lines using genomic and drug response characterizations. We will engineer resistance to clinical ALK inhibitors (alectinib and lorlatinib) and characterize the resistant cell lines to understand molecular pathways of ALK inhibitor resistance.
- Objective 2: Identify clinical drugs or drug combinations that can inhibit the growth of ALK inhibitor resistant cell lines. We propose to identify drugs and/or combination of drugs that can kill resistant EML4-ALK cell lines using a high throughout phenotypic screening platform.
- Objective 3: Develop novel therapies for ALK+ NSCLC. We propose to develop novel compounds that can degrade the EML4-ALK protein within the cell. Once developed, we will evaluate in our EML4-ALK cell lines, and we will compare the resistance pathways of our novel compounds to traditional ALK inhibitors. Our goal is to develop broad-spectrum ALK degraders that can degrade existing resistant mutations.
Developing Project: The ALK immune environment
Project Leader:
- Kiran Lagisetty, M.D., Ph.D.
The focus of this aspect of the initiative is to determine the immunologic mechanisms by which ALK positive tumors evade traditional immune based therapies. We have been developing strategies for the collection of primary untreated lung cancer and peripheral blood mononuclear cells (PBMC), confirmed ALK-fusion patient tissue sections for multiplex IHC, and bulk RNA sequencing to identify immune cell subsets associated ALK tumor development.
Project Objectives:
- Objective 1: Characterize the immune microenvironment in ALK NSCLC utilizing patient derived tumor tissue and identify significant differences immune populations (e.g. T-cells, neutrophils, macrophages, dendritic cells, cancer associated fibroblasts, etc.), from non-ALK mutation-driven NSCLC and non-mutation-driven NSCLC.
- Objective 2: Characterize the cytokine profiles of PBMCs in ALK NSCLC patients longitudinally, as treatments and clinical status evolve. Furthermore, we will characterize the immunologic states of various immune cells such as T-cells and circulating monocytes and neutrophils. We will compare this to PBMCs of healthy patient donors.
- Objective 3: Evaluate efficacy of current and emerging immunotherapies in ex vivo tumor organoid models and determine mechanisms of immunologic mediated tumor resistance.
Developing Project: Circulating Tumor Cells (CTCs) Analysis
Project Leader:
- Sunitha Nagrath, Ph.D.
We are currently in the process of developing technologies to isolate, enrich, and expand circulating tumor cells from patient-derived samples in order to characterize tumorigenesis and metastatic potential.
Project Objectives:
- Objective 1: Characterize the number and phenotype of circulating tumor cells in ALK NSCLC patients longitudinally, as treatments and clinical status evolve.
- Objective 2: Apply multiomic approaches to characterize the CTCs, including genomic and transcriptomic analysis. Developing workflows to characterize the CTCs at single cell level to identify heterogeneity and how it lends to treatment resistance.
- Objective 3: Expand the CTC population under conditions that preserve the phenotype and heterogeneity to fully characterize drug sensitivity and resistance.
- Objective 4: Use expanded CTCs optimize the protocols for further generation of xenograft models.
- strong>Objective 5: Understand the brain metastatic potential of CTCs.
Developing Project: Identification of Novel ALK Inhibitors
Project Leader:
- Peter Toogood, Ph.D.
Our goal is to identify novel ligands and inhibitors of ALK as lead compounds for the development of new ALK-targeted therapies. To this end, we are pursuing two parallel approaches. In the first approach we are using various screening technologies to identify ligands for purified human ALK protein. These screens are designed to preferentially identify allosteric ligands, i.e. compounds that bind at sites outside the known ATP binding pocket. These ligands have the potential to act independently as inhibitors of ALK function. In addition, allosteric ALK ligands maybe incorporated into heterobifunctional PROTAC molecules for targeted ALK degradation. Our second approach to the identification of ALK inhibitors uses structure-based de novo design to identify entirely original ligand structures, which we then prepare and test for inhibition of ALK. We are employing two different design platforms to identity either allosteric or ATP site ligands. Through these combined efforts we expect to discover new compounds that inhibit ALK with the potential to overcome resistance to approved ALK-targeted drugs.
Project Objectives:
- Objective 1: Identify allosteric ALK ligands by screening.
- Objective 2: Identify ALK ligands by de novo design.